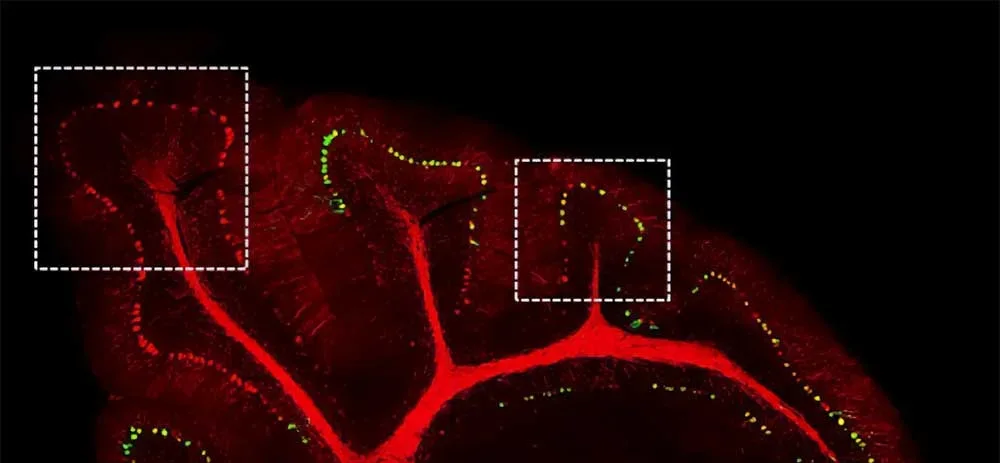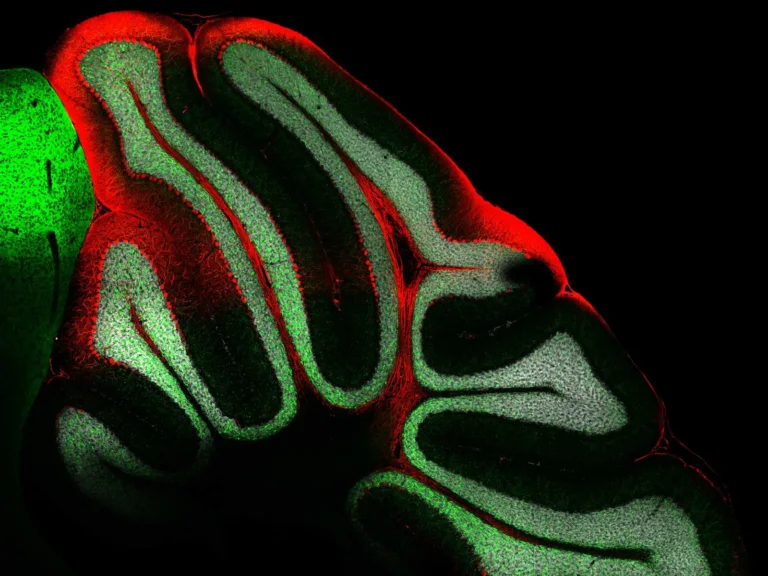
These are signatures easily observable in neurodegenerative diseases but hard to isolate, constraining discovery of the proteome and mechanisms underlying disease.
Synaptic plasticity, multicellular interactions, and regional brain signaling involve complex protein functions. Conventional protein assays are unable to systematically isolate proteomes from these subcellular and nanoscopic regions, leaving many mechanistic questions unanswered.

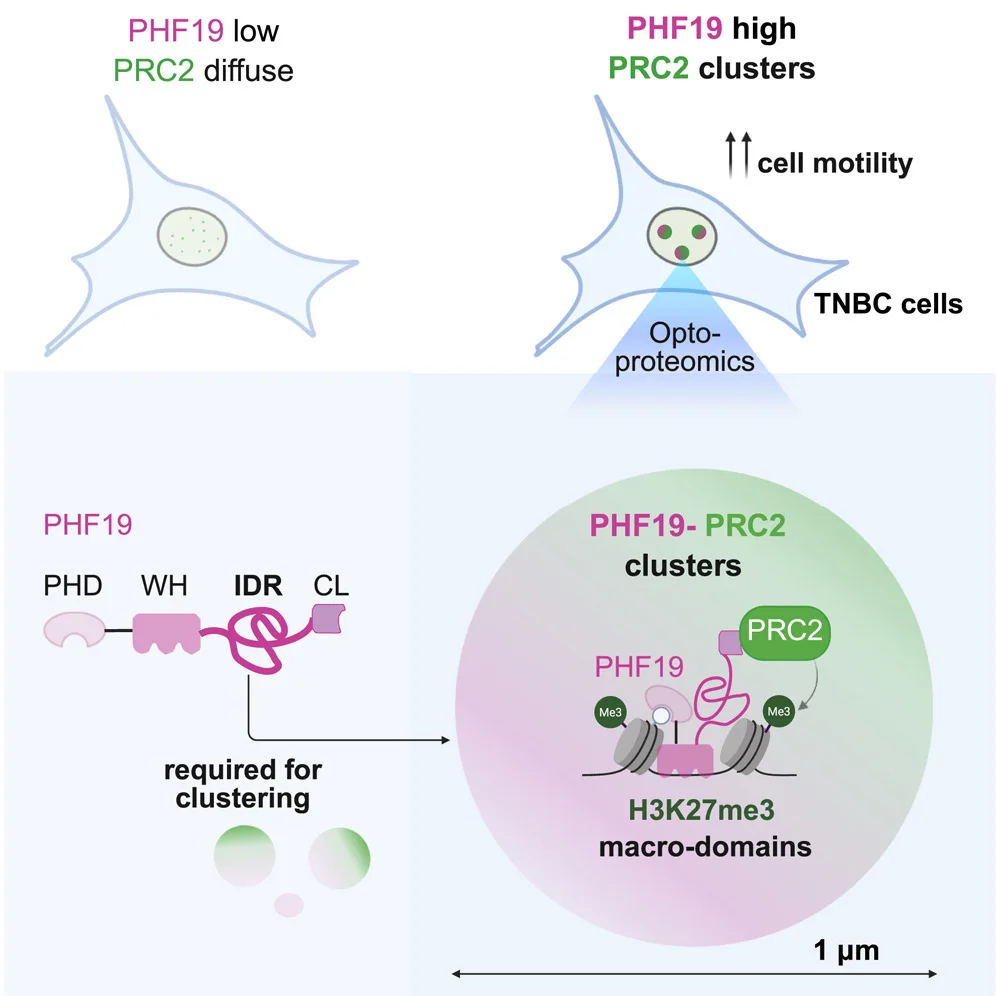
Microscoop combines optical targeting with proximity-based protein tagging, allowing researchers to illuminate and identify proteins within defined nanometer-scale regions of interest.
By coupling optics and proteomics, proteins can be defined not just by presence, but by “where” they operate in the cell and how those local interactions change under perturbation.
Microscoop enables researchers to: