Understanding the interplay between mitochondria and lipid droplets is critical to decoding the molecular underpinnings of metabolic disorders such as non-alcoholic fatty liver disease (NAFLD). Azad Gucwa presented a webinar on the preprint of our study findings, highlighting Syncell’s groundbreaking spatial proteomics workflow that maps proteins at the subcellular interface with nanoscale precision and identified novel proteins with therapeutic potential.
NAFLD has reached epidemic proportions, affecting an estimated 100 million individuals in the U.S. alone. Its prevalence is rising sharply among children and is closely associated with obesity and metabolic dysfunction. As the disease progresses from benign fatty liver to non-alcoholic steatohepatitis (NASH), cirrhosis, and even hepatocellular carcinoma, the need for mechanistic insights and targeted interventions has become urgent.
Central to this pathophysiology is the dynamic relationship between mitochondria and lipid droplets — two organelles intimately involved in lipid metabolism. Mitochondria drive fatty acid oxidation, while lipid droplets store fatty acids and coordinate their availability. Disruption at this interface, also known as the peridroplet mitochondrial (PDM) interface, can impair mitochondrial function, alter organelle morphology, and shift lipid homeostasis.
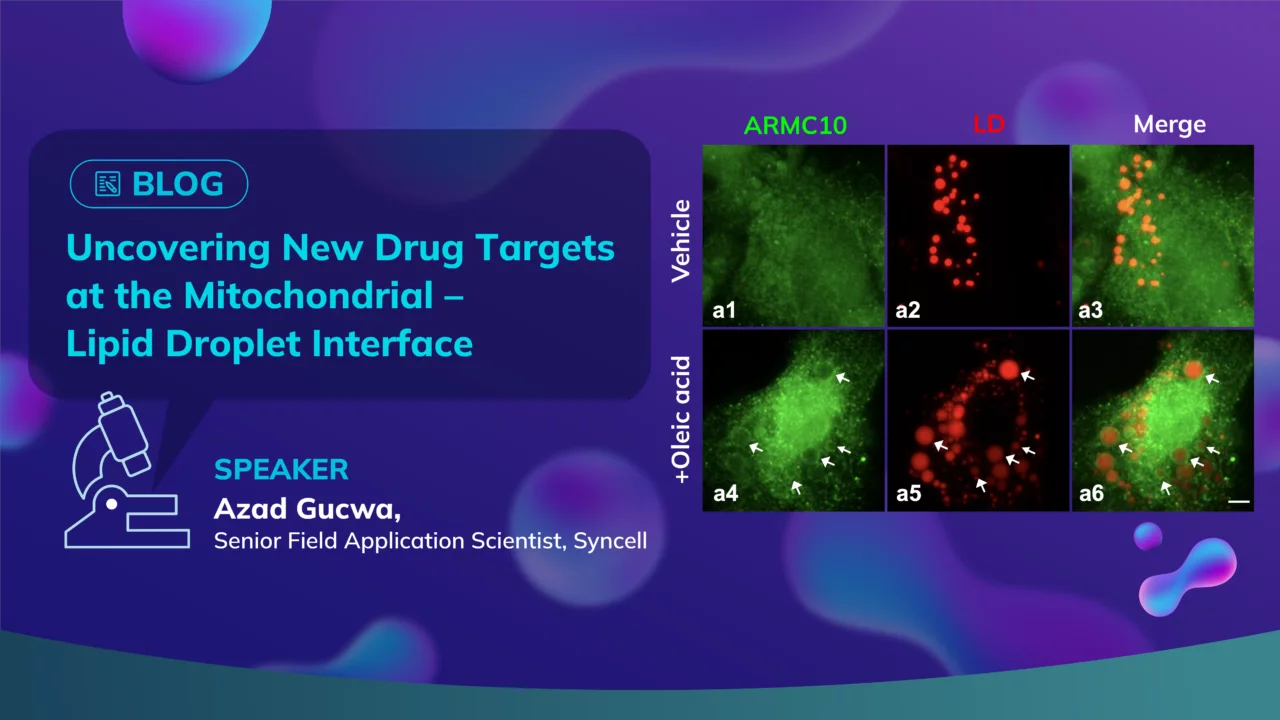
To investigate this interface in detail, the team used hepatocyte-derived HepG2 cells treated with oleic acid to promote lipid droplet formation. Using Tom20 and perilipin-2 as markers for mitochondria and lipid droplets, respectively, they leveraged an advanced microscope platform to selectively photolabel and biotinylate proteins specifically at the contact sites. These labeled proteins were then identified via mass spectrometry.
The results were striking. From over 2,200 proteins enriched at the PDM interface, 75 were known lipid droplet-associated and 261 were mitochondrial — validating the approach. Even more compellingly, the team uncovered 14 previously unannotated proteins that had no known associations with either organelle but were highly enriched at the interface. These novel proteins may represent uncharted components of lipid regulation.
A subset of five candidate proteins stood out due to their predicted amphipathic helices — a structural feature enabling membrane association. Upon further imaging and validation, all five showed strong localization to lipid droplets, particularly after oleic acid treatment, suggesting they are actively recruited in response to lipid accumulation.
Among these, FHL3 (Four and a Half LIM Domains 3) emerged as a protein of interest. Not only did FHL3 localize robustly to the PDM interface, but siRNA-mediated knockdown of FHL3 also significantly reduced the number of lipid droplets and mitochondria contact sites. This was accompanied by mitochondrial elongation—a morphology typically associated with the regulation of fatty acid oxidation—suggesting a potential role for FHL3 in lipid metabolism.
These discoveries were only possible through the precision of Syncell’s unbiased spatial proteomics Microscoop technology. By targeting nanoscale regions within cells and identifying proteins in a hypothesis-free manner, the platform allows researchers to map complex subcellular environments that traditional methods overlook.
The implications are broad: this workflow not only deepens our understanding of metabolic disease mechanisms but also delivers a new class of candidate drug targets, such as FHL3, for future investigation.
This study demonstrates that marrying spatial resolution with proteomics enables powerful insight into cellular function and dysfunction. As the field of drug discovery continues to evolve, such integrative approaches will be key to uncovering novel targets and developing more precise therapies. You can watch the on-demand webinar here.